Statistics: principal components analysis
Principal components analysis (PCA) is an incredibly powerful statistical tool, but can be daunting to the uninitiated. Most discussions of PCA begin with eigenvectors and orthogonal linear transformation and covariance matrices. While these are cool, it is possible to interpret a PCA without a deep understanding of p-dimensional ellipsoids.
In untargeted metabolomics, PCA is most often used to plot samples by their chemical similarity. Samples with similar metabolic profiles will cluster together, while less similar samples will be farther apart. PCA can simplify a dataset with thousands of metabolites into a handful of essential features, called principal components, and use these features to differentiate between samples.
Here are some steps to use when interpreting a PCA:
1. Is my PCA useful?
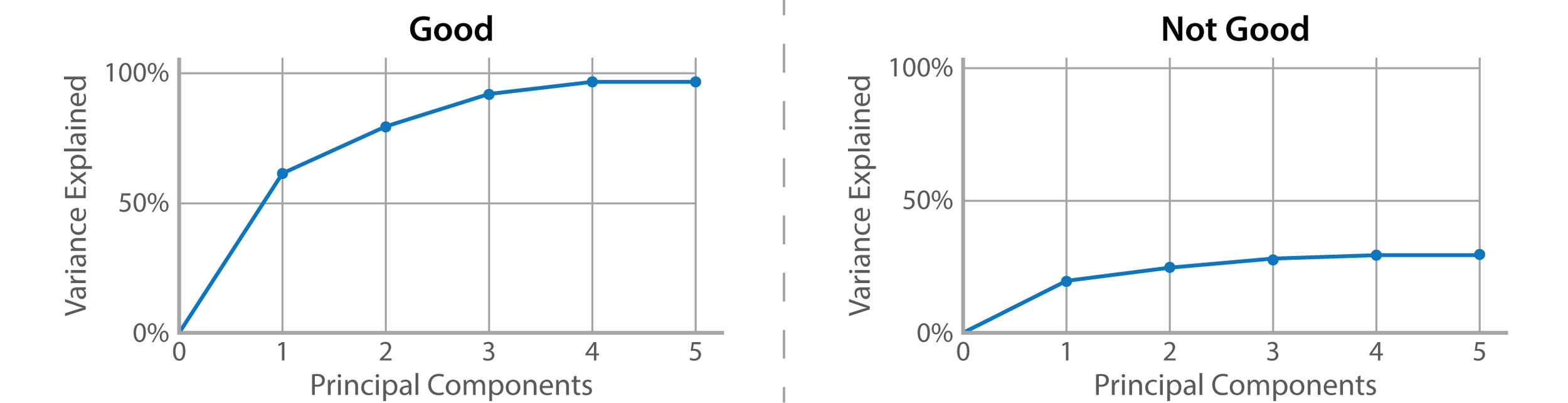
One of the first things to check when you’re looking at a PCA plot is how much variance is explained by each principal component. This metric tells you how well a principal component summarizes your data. Principal components (PCs) are ranked by how much variance they explain, so the first principal component (PC1) will always have a higher explained variance than PC2, PC3, etc. You want each principal component to explain a large amount of variance. If your explained variance is low, it means that your PCA isn’t useful.
2. Are my groups different?
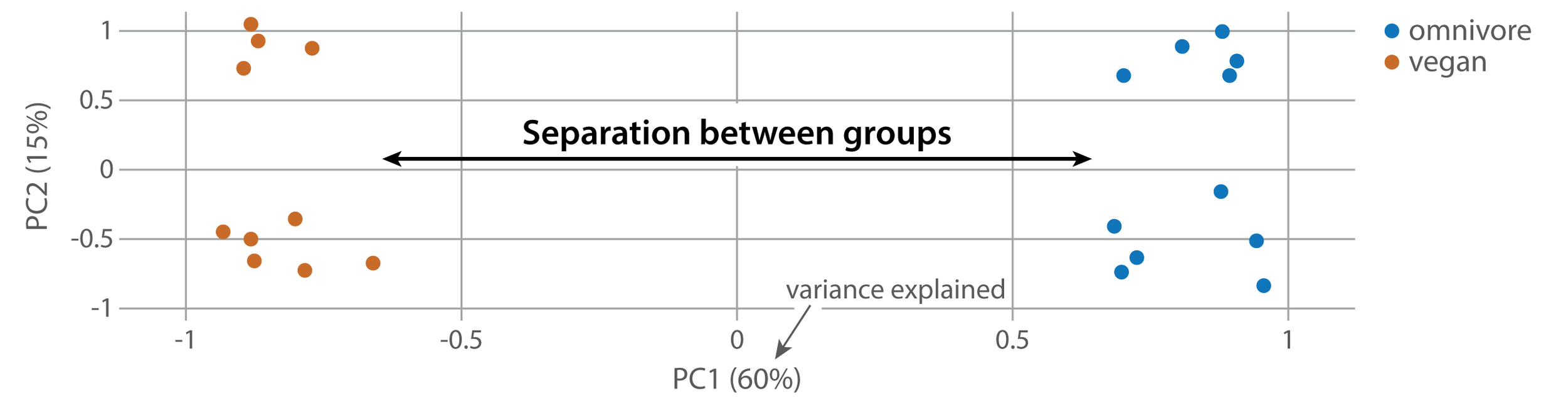
If you’re doing an experiment comparing two or more groups, ideally those groups will have different metabolic profiles. Thus, you’d expect samples from each group to cluster together and separate from samples in the other group. Here’s a lovely example of this:
Here, vegan and omnivore samples cluster on either side of the x axis (PC1). If your groups don’t cluster together, that indicates that there wasn’t a large metabolic difference between your groups.
That said, if your groups aren’t separating along PC1, don’t despair. It’s possible that a different principal component (say, PC4 or PC5) does separate your groups. This indicates that while there are differences between your groups, they aren’t the main drivers of metabolic difference in your experiment.*
My groups don’t separate, now what?
Sometimes you’ll find that your experimental groups don’t separate on any of the PCs. The first thing to check for in this case is whether there is any separation between your samples, even if it’s not within your experimental groups. If your samples seem randomly sorted into clusters, something is creating metabolic differences that aren’t related to your experimental design. If all your samples are randomly distributed without any clusters, there aren’t large metabolic differences within your experiment. PCA tends to be good at identifying large overall differences in metabolism but may fail when metabolic differences are subtle. In this case, check out results from ANOVAs, volcano plots, or machine learning models to see if they were able to identify changes in individual metabolites.
3. Are there any differences within my groups?
In the plot above, there is good separation between omnivore and vegetarian groups along PC1. However, the picture gets a little more complicated looking along the y axis (PC2). Here, it looks like both omnivore and vegan samples separate into two sub-groups. This would be a great opportunity to dig further into your metadata to figure out why those samples may be different from one another.
4. Are there any outliers?
In a PCA plot, poor-quality samples can be very obvious. If all your samples cluster together except for one, that’s a good indication that you need to go back and figure out why the metabolic profile of that sample is so different.
5. What is driving these differences?
While it’s nice to know that your groups are metabolically different, what you really want to know is which metabolites separate the groups. For this, you can use feature importance - a metric of how much each metabolite contributes to each principal component. The larger the feature importance, the more important that metabolite is for separating your groups.
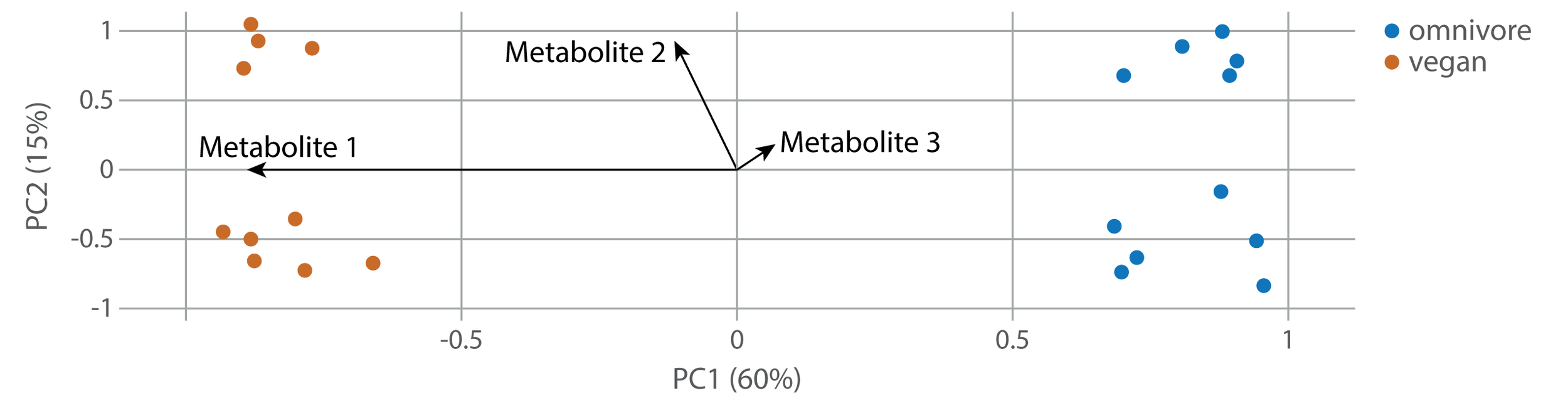
Feature importance is the basis for one of the most common visualizations of PCA: the biplot. A biplot is a PCA plot with vectors drawn for single metabolites that indicate their feature importance along both PCs.
In the biplot above, Metabolite 1 is very important for PC1 (separating omnivores and vegans), but doesn’t make any difference to PC2. Metabolite 2 is more important at explaining differences in PC2 than in PC1, and Metabolite 3 isn’t really making much of a difference. This is what you would see if you created a boxplot of each of these metabolites:
The Ometa Flow Principal Components dashboard allows you to sort and filter metabolites based on their feature importance for each PC. You can then click on a component to draw the biplot vector for that specific metabolite.
PCA
While PCA is the most popular method to separate data into its principal components, there are many other algorithms that can perform similar tasks. In Ometa Flow’s statistics processing workflow, we automatically run PCA, singular value decomposition (SVD), independent component analysis (ICA), and sparse principal component analysis (sPCA) and output the results of all four algorithms so you can compare between them. Check out the workflow documentation for more information on each of these algorithms.
*An example of when this might happen: you’re collecting samples from two patient groups and collection happens either morning or evening. It’s possible that the time of collection significantly changes metabolism and ends up separating your samples along PC1. However, you may still be able to see separation between patient groups in other PCs - they’ll just be smaller than the changes caused by time of collection.
Further Reading
Greenacre, M. et al. Principal component analysis. Nat. Rev. Methods Primer 2, 1–21 (2022).
A great primer on PCA (but unfortunately behind a paywall).
Jolliffe, I. T. & Cadima, J. Principal component analysis: a review and recent developments. Philos. Transact. A Math. Phys. Eng. Sci. 374, 20150202 (2016).
A more math-heavy explanation of PCA.
Have more questions? Contact us.